Projects
My laboratory is interested in the study of mobile element related genetic variation. The Alu family of mobile elements comprise approximately 10% of primate genomes and are responsible for a number of different genetic disorders (see our recent reviews on mobile elements see Batzer and Deininger, 2002; Callinan and Batzer 2006; Cordaux and Batzer, 2009; Deininger and Batzer, 2002: Deininger et al., 2003). These elements are one type of L1 element dependent retrotransposon that is specific to the Primate order. A third composite retrotransposon termed SVA is restricted to hominids and also dependent on L1 elements for mobilization (Damert et al., 2009; Wang et al., 2005). Collectively these retrotransposons make up a significant proportion of primate genomes both in terms of copy number and overall mass as shown in the adjacent figure. Mobile elements play a significant role in the generation of genomic diversity through a variety of processes such as insertional mutagenesis (reviewed in Deininger and Batzer 1999; Batzer and Deininger 2002; Cordaux and Batzer 2009; Konkel and Batzer, 2010), transduction (Xing et al., 2006), recombination (Sen et al., 2006) and double strand break repair (Sen et al., 2007).
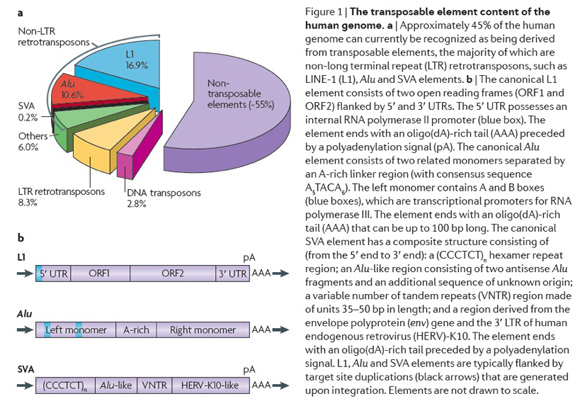
Initial research on retrotransposable elements demonstrated that, although these elements exist at a very high copy number, individual subfamilies of the elements of different genetic ages exist within the genome. Members of the recently integrated Alu, L1 and SVA subfamilies are restricted to specific parts of the primate lineage including human lineage specific elements (reviewed in Batzer and Deininger 2002; Cordaux and Batzer, 2009). Many of these "young" mobile elements have inserted so recently within the primate lineage that individuals can be polymorphic for the presence or absence of a mobile element at a particular chromosomal location. Mobile element insertion polymorphisms offer two important advantages over other nuclear based polymorphisms for population genetics and phylogenetic studies (reviewed in Ray et al., 2006). First, the presence of a mobile element insertion represents identity by descent, since the probability that two different young mobile elements would integrate independently in the same chromosomal location is negligible. Second, the ancestral state of each mobile element insertion polymorphism is known to be the absence of the mobile element, which can be used to unambiguously root trees. The insertion of mobile elements into the genome represents a novel class of nuclear markers for the study of population genetics and phylogenetic relationships.
The research within our laboratory is focused around the characterization of mobile element based genetic variation. There are currently several research foci within the laboratory.
Genomic Impact
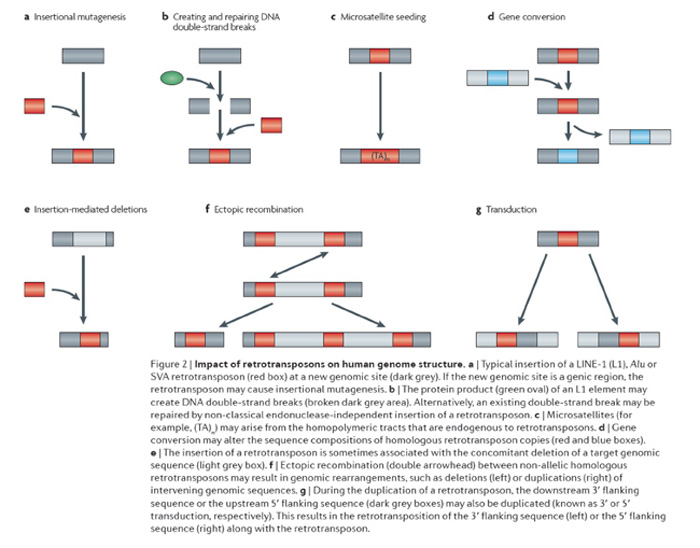
We are interested in determining the diverse roles that mobile elements have played in shaping the architecture of mammalian genomes. Currently we are in the process of identifying and characterizing new mobile elements throughout the primate order (de Koning et al., 2011; 1000 Genomes Consortium 2010; Gibbs et al., 2007; Han et al., 2005, 2007; Locke et al., 2011; Wang et al., 2005) and in other mammals (Gentles et al., 2007; Gu et al., 2007; Mikkelsen et al. 2007) to gain insight into the amplification dynamics of these elements. We are also interested in defining the contributions that mobile elements make to structural variation in genomes (Mills et al., 2011). A schematic diagram depicting many of the ways mobile elements contribute to structural genomic variation is shown in the adjacent figure (reviewed in Cordaux and Batzer, 2009). We have focused on the impact that mobile elements have had upon genomic diversity through the creation of deletions during mobilization (Callinan et al., 2005; Han et al., 2005), their involvement in recombination (Sen et al., 2006), in double strand break repair (Morrish et al., 2002; Sen et al., 2007) and in non-traditional insertion mechanisms (Meyer et al., 2010; Srikanta et al., 2009a,b). We are also studying the role that these elements play in the generation of genes and gene families through transduction (Gibbs et al., 2007; Xing et al., 2006, 2009) and how they contribute to the creation of new regulatory networks (Cordaux et al., 2006; Gentles et al., 2007; Mikkelsen et al., 2007).
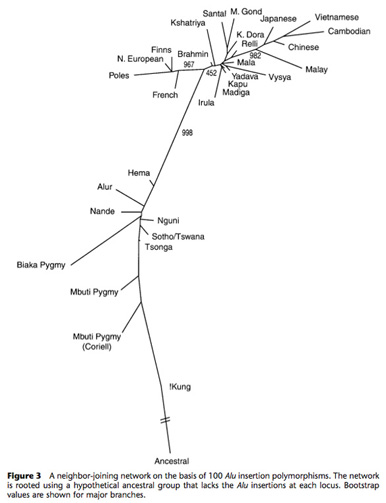
Human Population Genetics
We are also examining the levels of human genomic variation associated with mobile elements that are unique to the human lineage in collaboration with Drs. Lynn Jorde and Alan Rogers at the University of Utah (Bamshad et al., 2003; Rogers et al., 2007; Watkins et al., 2003; Witherspoon et al., 2006, 2007, 2009, 2010; Xing et al., 2009) and with multiple laboratories throughout the world in as part of 1000 Genomes Consortium (2010). A tree of human population relationships derived from the analysis of 100 Alu insertion polymorphisms is shown in the adjacent figure. In addition, we are interested in determining the impact that new mobile element insertions have upon the accumulation of other types of genetic diversity, and how gene conversion between mobile elements alters levels of single nucleotide polymorphism within the human genome (Roy Engel et al., 2002; Salem et al., 2003b; Vincent et al., 2003; Witherspoon et al., 2009; 1000 Genomes Consortium, 2010).
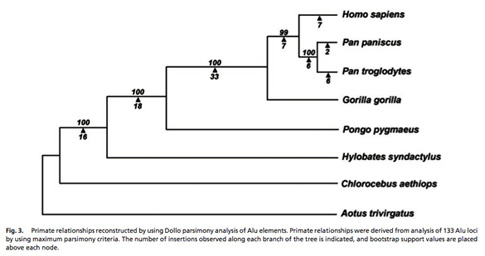
Primate Phylogenetic Relationships
We are utilizing a combination of computational approaches and PCR based assays to identify and characterize lineage specific Alu and L1 insertions in the Primate order in collaboration with Drs. Lynn Jorde (University of Utah), Oliver Ryder (San Diego Wild Animal Park), Todd Disotell (New York University), Christian Roos (German Primate Center), and Lucia Carbone (Oregon Health and Science University). Because of the known direction of polarity of mobile element insertions and absence insertion of homoplasy (across short evolutionary distances) these elements are a novel source of genetic variation for the study of primate phylogenetic relationships (Ray et al., 2006). We are in the process of determining the phylogenetic relationships between species throughout the primate order using mobile element insertion polymorphisms (Herke et al., 2007; Li et al., 2009; Ray and Batzer, 2005; Roos et al., 2011; Salem et al., 2003c; Xing et al., 2003, 2005, 2007). A tree of Hominid phylogenetic relationships derived from the analysis of mobile elements is shown in the adjacent figure.